Laboratory for Metabolomics
OBJECTIVES and ONGOING PROJECTS IN THE LAB
0. Overview
1. Regulation of inflammation and immunity by fatty acid metabolism
2. Creation of Lipidome Atlas to elucidate the biology of LipoQuality
3. Lipid metabolic interactions between symbiotic microorganisms and the host
Lipids constitute biological membranes and play a variety of roles as energy sources, signaling molecules, and their precursors. In addition, some lipids are biologically active as single molecules, while others are involved in the formation of membrane domains as molecular assemblies. Since lipids are extremely diverse molecules, the precise determination of different molecular species of lipid—a process we have termed “LipoQuality”, is important for understanding their functions in physiology and disease, and for discovering novel bioactive lipids that may have therapeutic benefits. In general, dysregulated lipid metabolism is associated with diseases such as obesity, atherosclerosis, stroke, hypertension and diabetes, and hence there is great interest in furthering our understanding of how they behave within complex biological systems. We aim to elucidate the mechanisms that create, regulate, and recognize lipid diversity as a spatio-temporal system by constructing a “Lipidome Atlas”, as well as to elucidate diseases caused by its disturbance.
In the history of science, the discovery of new substances and the elucidation of their molecular interactions have provided fundamental solutions to life phenomena and pathological conditions whose mechanisms have remained unknown, and have served as the basis for the formation and development of new scientific fields. The discovery of the significance of lipid molecules involved in various life phenomena and their diversity through non-targeted, highly accurate, and in-depth original technologies is expected to have a ripple effect on a wide range of life science fields, and contribute to medical science and a healthy and long-lived society based on scientific evidence.
1. Regulation of inflammation and immunity by fatty acid metabolism
Polyunsaturated fatty acids (PUFAs) exhibit a wide range of biological effects, many of which are mediated through the formation and actions of their bioactive metabolites. It is well appreciated that dietary PUFA balance affects inflammation and related diseases, and recent advances in mass spectrometry-based lipidomics technologies and their applications have revealed a potential link between PUFA metabolism and biological phenotypes. Our research is aimed at elucidating structure and function of endogenous lipid mediators that regulate inflammation and tissue homeostasis.
1-1. Omega-3 PUFA-derived lipid mediators
Omega-3 PUFAs such as eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) are widely held to have beneficial effects in many inflammatory diseases. Also, elevation in tissue levels of omega-3 PUFAs in fatty acid n-3 desaturase (fat-1) transgenic mice exhibits resistance to many inflammatory disease models. To date, we’ve developed targeted lipidomics system using LC-tripleQ MS to monitor fatty acid metabolites comprehensively, and uncovered the roles of omega-3 PUFA metabolites in controlling inflammatory responses. For example, 18-HEPE, which we identified as an active metabolite that inhibits heart failure (fibrosis of myocardial tissue), is also a metabolic precursor of the resolvin E series, an anti-inflammatory metabolite derived from EPA. In addition, 17,18-EpETE, which is generated by epoxidation of EPA, was found to have antiallergic activity, and a series of active metabolites originating from 17,18-EpETE, such as 12-OH-17,18-EpETE, were also identified. These are metabolic pathways unique to omega-3 PUFAs, in which the double bond at the n-3 position is oxidized, and we termed this pathway the "omega-3 cascade”. These endogenous mediators with potent anti-inflammatory property could lead to the development of novel therapeutics.

1-2. PUFA metabolism involved in the regulation of inflammation and allergy
Acute inflammation is an indispensable host response to foreign challenges or tissue injury. In healthy conditions, inflammatory processes are self-limiting and self-resolving, suggesting the existence of endogenous mechanisms for the control of inflammation and resolution. Comprehensive understanding of the cellular and molecular events of a well-orchestrated inflammatory response is required, and recent studies have uncovered the roles of endogenous lipid mediators derived from PUFAs in controlling the resolution of inflammation. Polyunsaturated fatty acid (PUFA)-derived mediators are formed by enzymatic oxidation through the action of cyclooxygenases (COX), lipoxygenases (LOX), and cytochrome P450 monooxygenases (CYP). By using LC-MS/MS-based mediator lipidomics, we provided new insights into the cellular and molecular mechanisms of inflammatory resolution, especially the functional roles of 12/15-LOX-expressing eosinophils and macrophages in controlling the resolution of inflammation.
Mediator lipidomics concerns the simultaneous and quantitative analysis of bioactive lipid mediators in biological systems. This technology could potentially identify the metabolic fingerprint of a disease for clinical diagnosis and treatment. Indeed, we revealed impaired 12/15-LOX activity in eosinophils isolated from allergic patients as compared to the healthy subjects. Also 12/15-LOX-deficient mice were more susceptible to illness of influenza virus infection. These results indicate the potential link and protective role of endogenous lipid mediators biosynthesized via the 12/15-LOX-mediated pathway. Moreover, endogenous lipid mediators with anti-inflammatory and tissue-protective actions could lead to the development of novel therapeutics for human disease when sustained inflammation is suspected as key components of pathogenesis.

1-3. Mechanism of the fatty acid quality management in specific tissues
Specific organs such as brain, retina, and testis were enriched in long-chain polyunsaturated fatty acids (LC-PUFA)-containing lipids including DHA-containing phospholipids. We have discovered the importance of lipid quality containing LC-PUFA in the establishment of a membrane environment that optimizes brain function, retinal photoreception, and spermatogenesis using acyl-CoA synthase 6-deficient mice. We hope to elucidate why specific fatty acids such as DHA are necessary for optimizing biological functions and localization analysis by mass spectrometry imaging.
2. Creation of Lipidome Atlas to elucidate the biology of LipoQuality
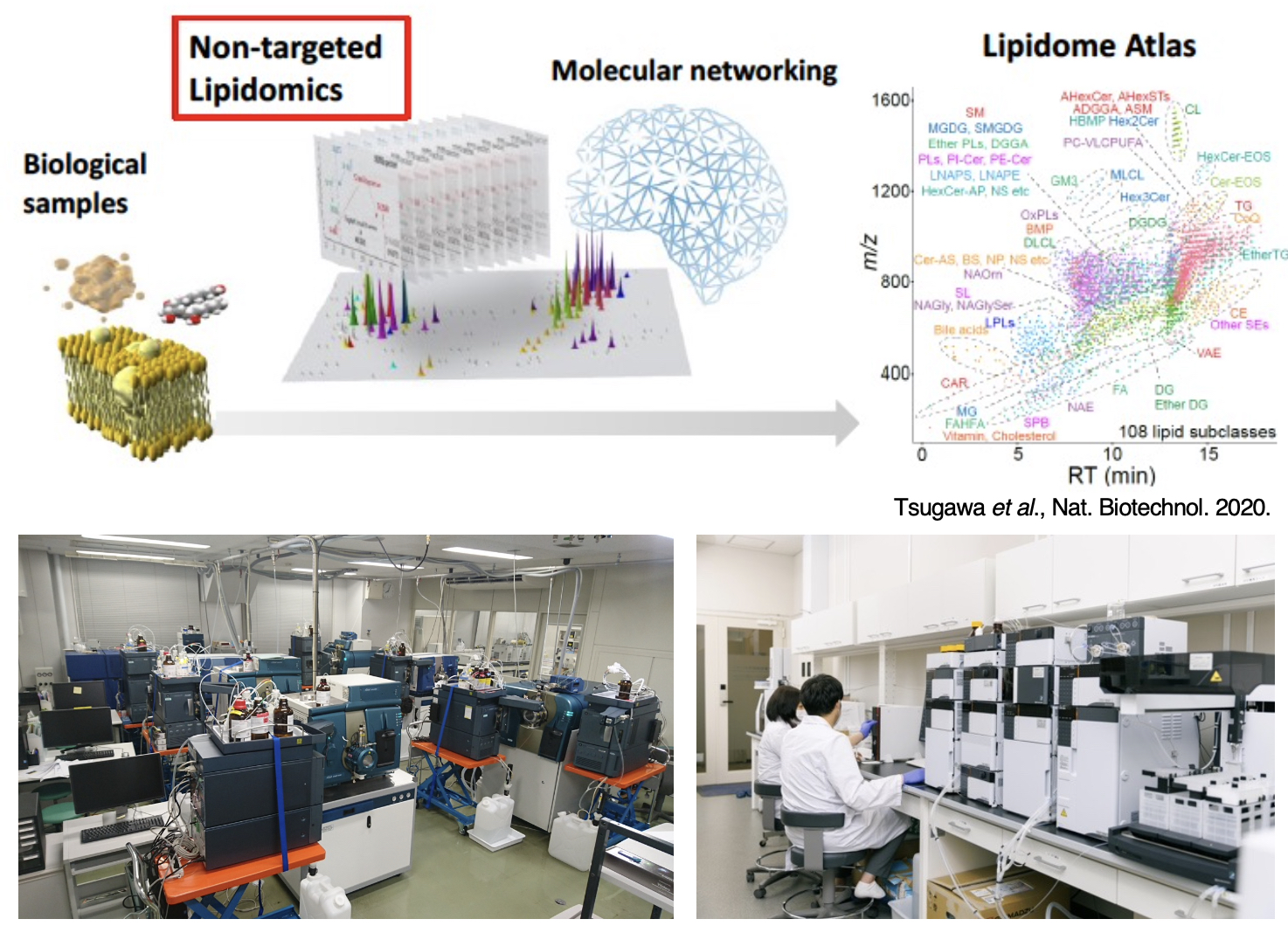
2-1. Development and application of non-targeted lipidomics
It is estimated that more than 100,000 types of lipid molecules are present in living organisms. We have successfully developed a non-targeted lipidomics platform using LC/Q-TOF MS with MS-DIAL4 software that derived a catalog of over 8,000 unique structures of 117 lipid class categories. We created a comprehensive database of lipid structures with their mass spectrum properties such as retention time, collision cross section, and mass fragmentation pattern to correctly characterize lipids. This non-targeted lipidomics platform is a powerful technology for visualizing lipid networks globally, and provides an opportunity for data-driven hypotheses to make the connection between lipid metabolism and biological phenotypes.

2-2. Mass spectrometry imaging and spatial multi-omics analysis
In order to uncover how lipid diversity is created in vivo and how its localization and metabolizing pathways are regulated, it is necessary to identify the transcripts and enzymes (including their activities, post-translational modifications) that are involved in the homeostasis of each lipid species. In the JST-ERATO Lipidome Atlas Project, we develop a cutting-edge technology of mass spectrometry-based tissue imaging to capture a global view of the diversity, distribution, and localization of lipids in living organisms. We combine spatial lipidomics to investigate the localization of lipids, innovative proteomics to understand lipid metabolic enzymes and modifications, and spatial transcriptomics to elucidate the factors of lipid localization. In this way, we visualize the effects of the local environment created by specific lipids on the dynamics and functions of multicellular systems.

2-3. Chemical biology for identification and quantification of lipid-related proteins
Lipid diversity in living systems is generated by the activity of various lipid-metabolizing enzymes present in the local environment. Since lipid-metabolizing enzymes can be activated by post-translational modifications such as phosphorylation or by binding of cofactors, it is important to capture the activity of lipid-metabolizing enzymes in addition to their expression levels in order to elucidate the mechanisms that create and regulate lipid diversity. In recent years, activity-based protein profiling (ABPP) has emerged as a novel method in the field of chemical biology that enables proteome-wide identification and quantification of the activity of proteins using activity-based probes that bind to hyper-reactive and functional amino acid hotspots such as active sites of enzymes. Our laboratory is using ABPP to search for enzymes that regulate the local lipid environment in tissues, and to discover novel regulatory mechanisms of lipid metabolism.
Protein lipidation such as palmitoylation and prenylation is a well-known post-translational modification that regulates the cellular localization and function of target proteins. On the other hand, lipidated proteins are generally difficult to analyze due to their unique chemical properties and low abundance, and thus it is expected that a lot of lipid modifications still remain undiscovered. Our laboratory has succeeded in comprehensively identifying lipid-modified proteins in cells using alkyne-tagged lipids and click chemistry. Utilizing those alkyne probes will be able to discover novel lipid modifications and elucidate their physiological significance.

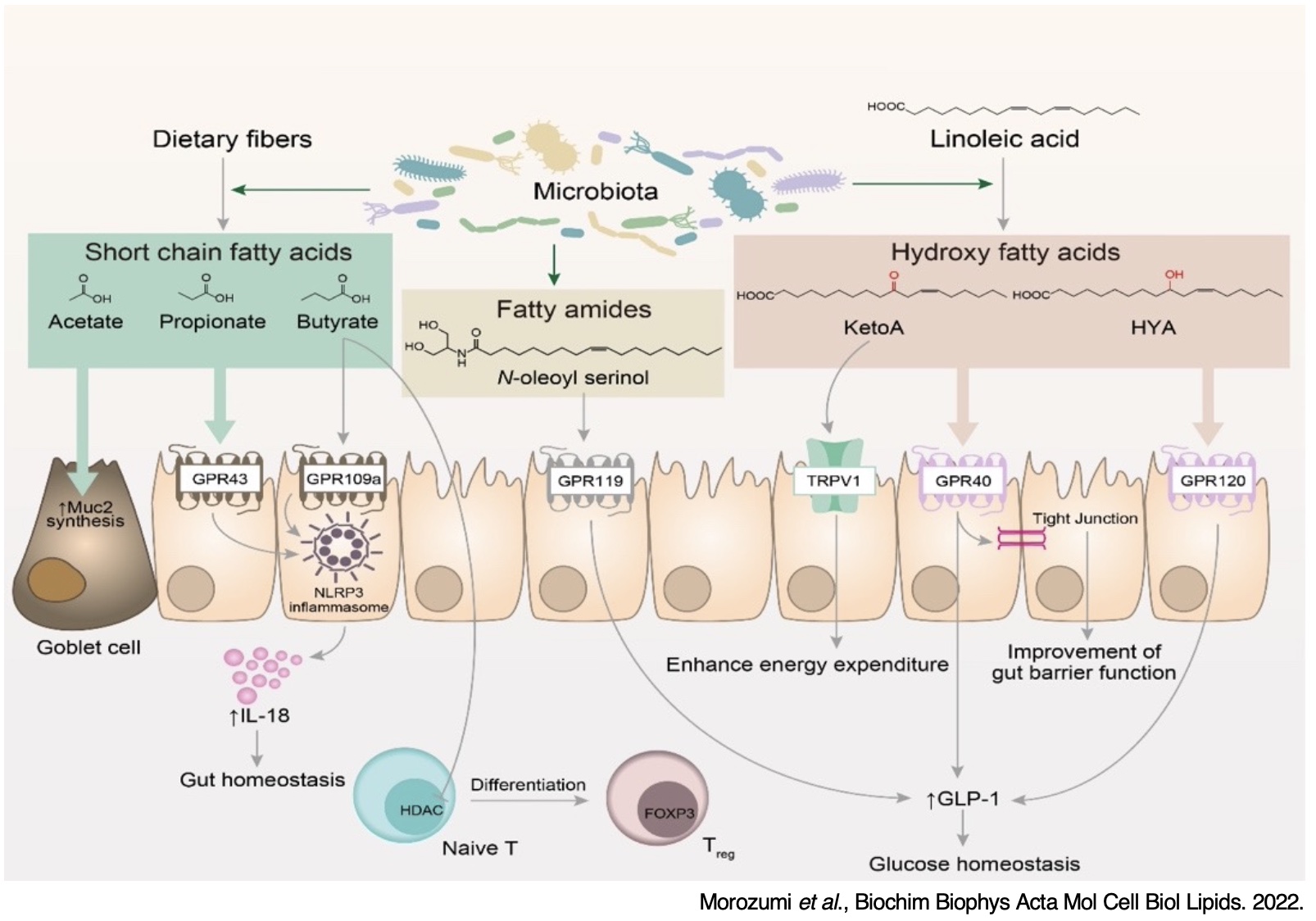
3. Lipid metabolic interactions between symbiotic microorganisms and the host
Intestinal flora exhibits their own metabolic systems, and the complex metabolic network created by the intestinal microbiota, dietary environment and host tissues remains to be resolved. Dysbiosis is closely related to various diseases such as inflammatory bowel disease, colorectal cancer, metabolic syndrome, and allergies, and is attracting attention as a new target for therapeutic intervention. In particular, the structures and functions of the gut microbial lipidome remain largely unknown. We have revealed that structure of lipid metabolites produced by intestinal bacteria, and these metabolites are transferred not only into the intestinal tract but also into host tissues. By combining untargeted mass spectrometry, which enables comprehensive analysis including unknown metabolites, and molecular spectrum networking technology, which supports structural estimation of unknown molecules, we aim to discover new functional metabolites in addition to clarifying the correlation between metabolites and the bacterial flora. We will elucidate through which bacteria the unique lipid metabolites are produced, and will clarify the spatial distribution in which lipid metabolites are produced and how they act on the host.