3. Development of technologies for next-generation trans-omics
We develop next-generation trans-omics technologies to integrate omic layers changing in different time scales for example comparatively fast omic layers (metabolome, metabolic flux, and phosphoproteome) and slower layers (epigenome, transcriptome, and expressing proteome) (Fig. 3). Furthermore, in collaboration with other laboratories, we incorporate large-scale data such as lipidome that have not been utilized in current trans-omics studies (Fig. 4). It is expected that incorporation of lipidome might allow trans-omic approaches to be applied to the analysis of a broader range of diseases and biological phenomena that cannot be handled with current trans-omics technologies.
Figure 3 | Integration of lipidome data with other omic data.
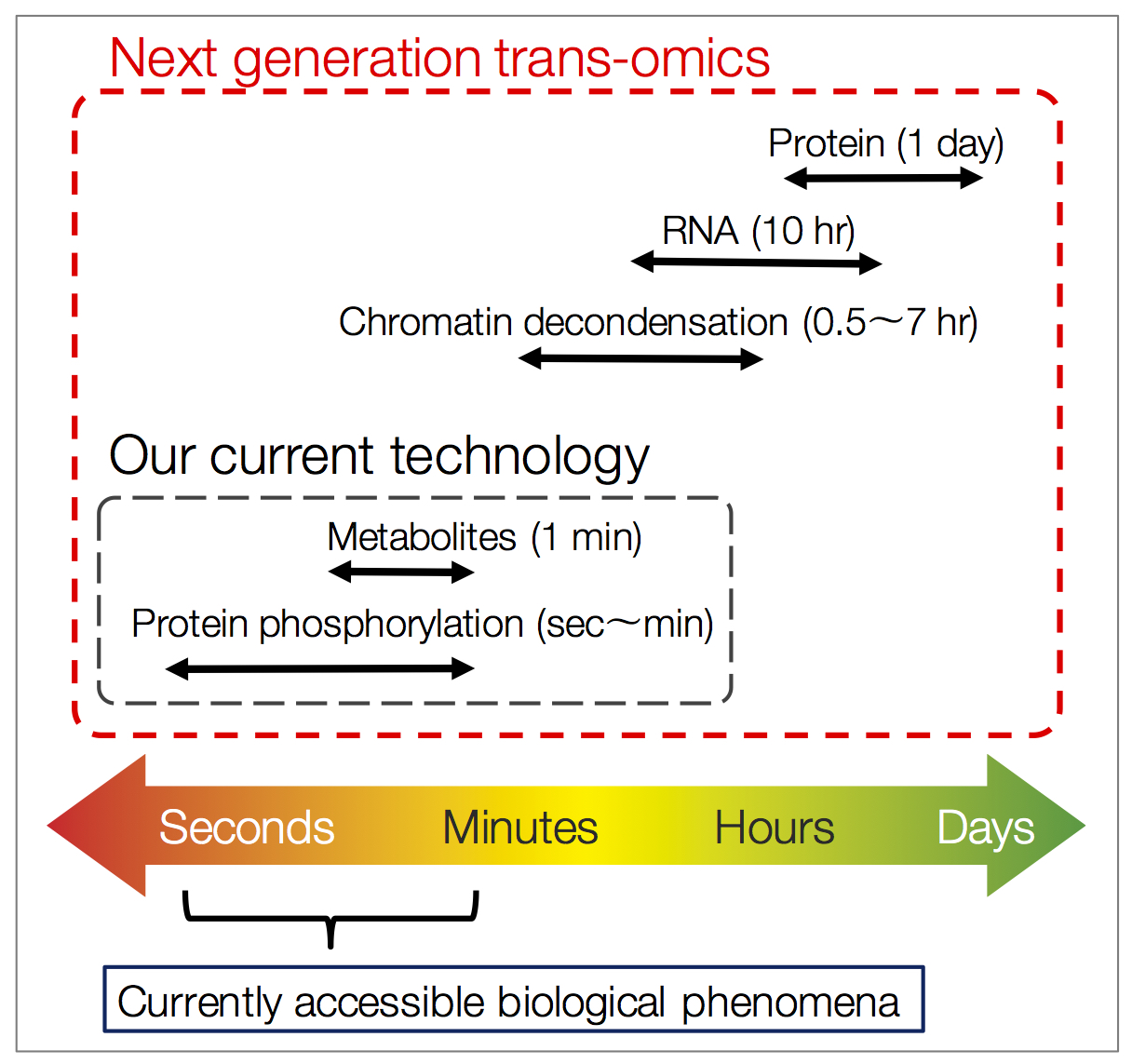
Currently, our trans-omics technologies cover integration of two ‘fast’ omic layers, metabolome and phosphoproteome.
We develop next generation trans-omics technologies to integrate also ‘slow’ gene expression-related layers so that one could apply trans-omic approaches to broader biological phenomena.
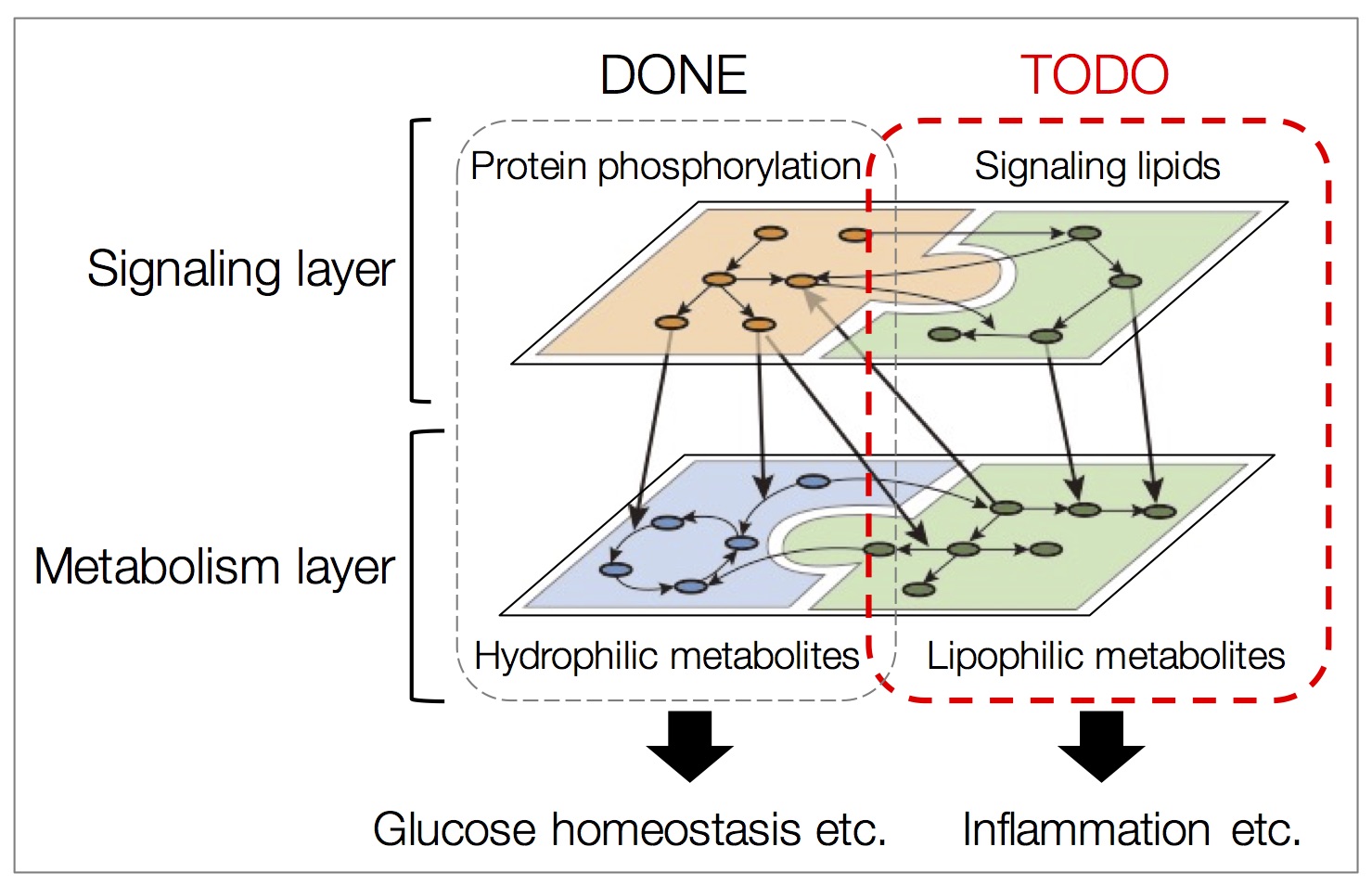
Figure 4 | Integration of lipidome data with other omic data.
Extending trans-omics, which is currently based on phosphorproteome and hydrophilic metabolites (left half), by integrating with lipidome data that constitute the signaling and metabolism layers (right half).
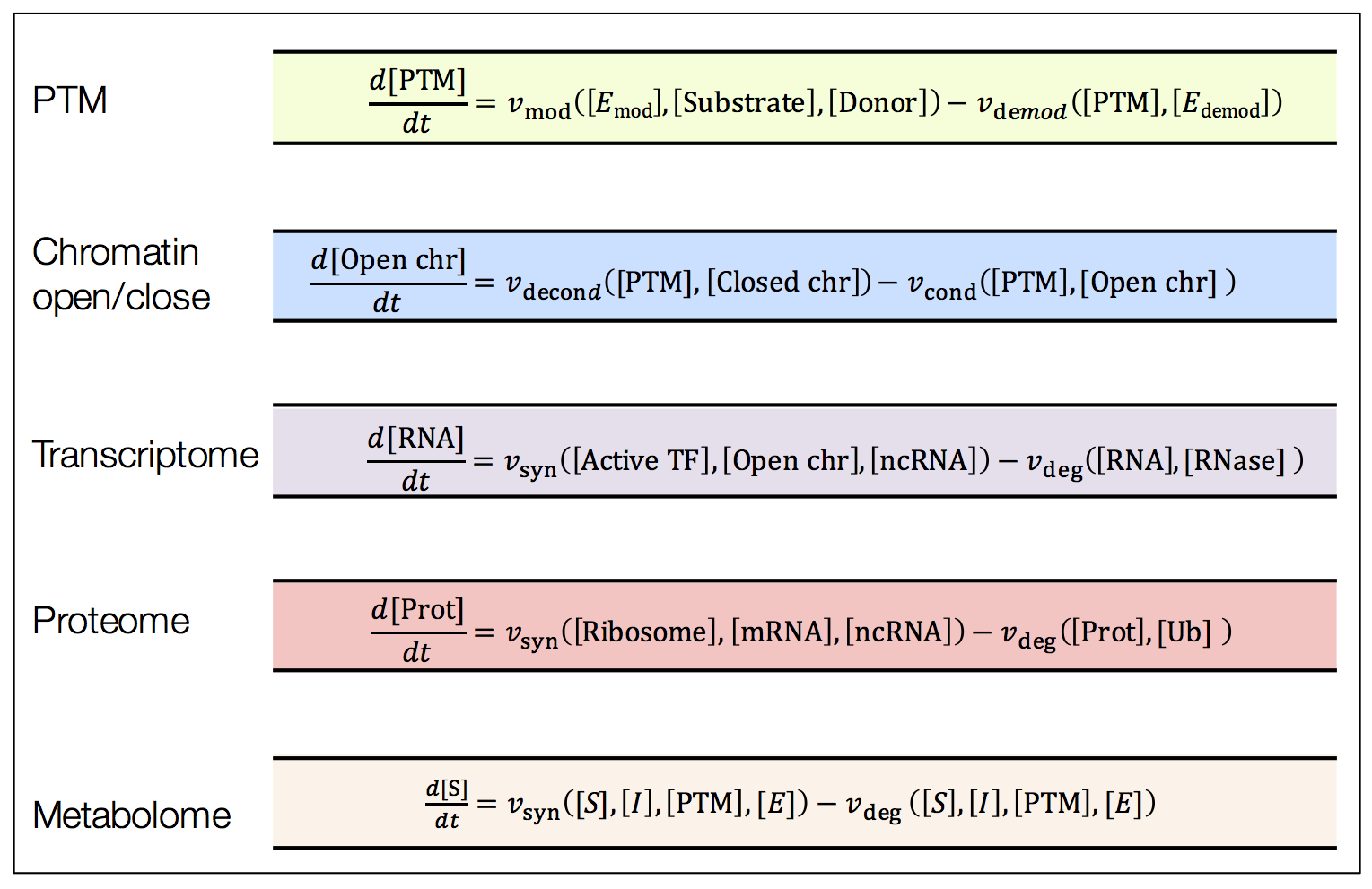
Figure 5 | Omic layers in rate.
Each reaction rate (terms represented by ‘v’) is a function of amounts of molecules that belong to same or different omic layers. Causalities across omic layers are reconstructed based on reaction kinetics.
|

.jpg)