Diverse forms of small RNA (sRNA) are found involved in numerous biological processes such as transcriptional and post-transcriptional regulation, epigenetic modification of chromatin, control of genome stability and cell-to-cell communication. Identification of important, functional sRNA species is currently hampered by the lack of reliable and sensitive methods to isolate and characterize them. Many native sRNAs are difficult to detect and quantify due to their unique molecular features, including short length, stable secondary structure, low abundance, lack of recognizable features like poly-A tails, presence of isoforms (for example isomiRs) and multiple precursor/intermediate forms.
In the research published in Nature Protocols, the scientists at RIKEN Center for Integrative Medical Sciences in Japan and IFOM-The FIRC Institute of Molecular Oncology in Italy developed a method termed Target-Enrichment of small RNAs (TEsR). The method enables targeted sequencing of rare sRNAs and diverse precursor and mature forms of sRNAs not detectable by current standard sRNA sequencing technologies. TEsR procedure is fully customisable, from the amplification of full-length sRNA molecules, production of biotinylated RNA probes, hybridization to one or multiple targeted RNAs, removal of non-targeted sRNAs, to high-throughput sequencing. By this approach, target sRNAs can be enriched by a factor of 500-30,000 while maintaining strand specificity. TEsR enriches for sRNAs irrespective of length or different molecular features, such as the presence or absence of a 5’-cap or of secondary structures or abundance levels. Moreover, TEsR allows the detection of the complete sequence (including sequence variants, 5’ and 3’ ends) of precursors, intermediate and mature forms, in a quantitative manner. The method can be applied to a wide range of RNA samples from microbes, plants and animals.
[caption id="attachment_2744" align="alignright" width="695"]
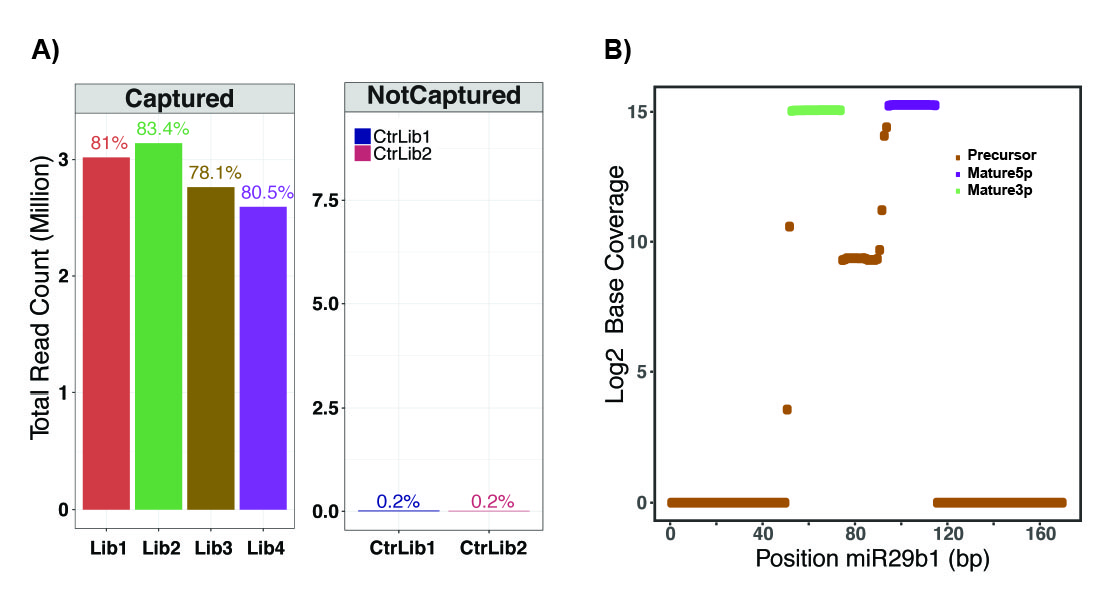 A) In TEsR, by capturing sRNAs of interest, ~ 80% of resulting sequence information is relevant to the target sRNAs (four replicates), as opposed to only ~ 0.2 % in a standard sequencing protocol (two replicates).
A) In TEsR, by capturing sRNAs of interest, ~ 80% of resulting sequence information is relevant to the target sRNAs (four replicates), as opposed to only ~ 0.2 % in a standard sequencing protocol (two replicates).
B) The resulting data were mapped at single-nucleotide resolution for both mature and precursor isoforms of target small RNAs (miR29b1), enabling accurate annotation and quantification. [/caption]
Piero Carninci of the RIKEN Center for Integrative Medical Sciences says, “We have developed an important method that can help to address the fundamental but challenging questions in small RNA research: whether or not (or in which conditions) the sRNA of interest exists; how many sRNA isoforms there are and what are their lengths, sequences and quantities; what are the precursors and intermediates of the mature sRNA; and how the expression levels of rare sRNAs change between different biological conditions. We believe the TEsR method will fill an important niche in the current technologies to study the cellular transcriptome and activity of the genome.”
Fabrizio d’Adda di Fagagna of IFOM and co-corresponding author on this publication, commented “This is the latest of the collaborative publications between our two research groups and provides further evidence of the great synergic potential between our two Institutes.”
Reference:
Nguyen, Q., Aguado, J., Iannelli, F., Suzuki, A.M., Rossiello, F., d'Adda di Fagagna, F., and Carninci, P. (2018). Target-enrichment sequencing for detailed characterization of small RNAs. Nature Protocols 13, 768.
https://www.nature.com/articles/nprot.2018.001
Related link:
article at RIKEN website
http://www.riken.jp/en/pr/topics/2018/20180419_1/
