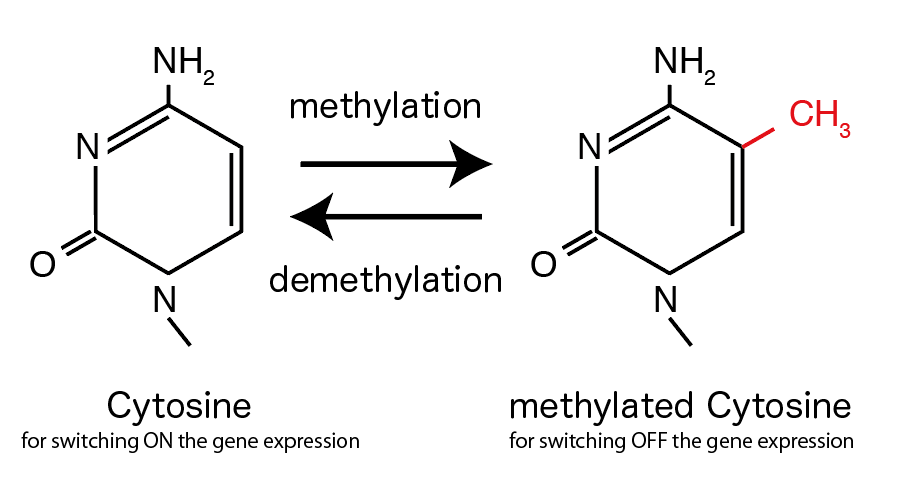
While human genome is considered to have ~20,000 genes, only a part of them is working in a single cell (Usually a half of them is expressed). In a cell, only the genes that are necessary for its survival and/or responsible for its function are active, and others are switched off. Cells have various mechanisms for switching ON and OFF the gene expression, and DNA methylation change is one of such mechanisms. DNA methylation/demethylation mechanism itself has been studied for many years, but how cell regulates the mechanism has remained to be solved.
Focusing on the extensive change of DNA methylation status upon blood cell differentiation, we showed that a protein named RUNX1 is regulating demethylation in this process. RUNX1 is a transcription factor (TF) known to bind to a specific DNA sequence to control gene expression in blood cells. Further analysis revealed that the DNA bound RUNX1 recruits demethylation enzymes to its binding site.
While more than 2,000 TFs are known to work in cells, very few TFs are reported to be involved in DNA methylation or demethylation. So, in the next step, we decided to comprehensively explore such TFs. For this purpose, we combined several methods to establish a novel versatile screening system to identify TFs that induce site-directed DNA methylation/demethylation. Using this system, we illustrated more than half (eight) out of target 15 TFs were novel DNA demethylation-inducing TFs.
This system will contribute to figure out the whole picture of relationship between TFs and DNA methylation change in the cell. Moreover, abnormal DNA demethylation is associated with several serious diseases such as cancers. Understanding the molecular mechanism how cell regulates DNA demethylation will provide a clue for better diagnosis and therapies for these diseases.