Projects
eQTL project: Integration of genetic information into immune functions
Many disease susceptibility variants have been identified by genome wide association study (GWAS). Germline genet-ic variations provide us with evidence into the causal relationship of an observed phenomenon and its pathogenesis. In this regard, the majority of GWAS risk variants have been reported to locate in the non-coding regions on the chromosome and function as an expression-quantitative trait locus (e-QTL), regulating the expression levels of genes. Therefore, by integrating genomic in-formation, qualitative and quantitative analyses of transcriptomes together with cell-specific epigenomes, we will better understand the causal pathogenic components of immune cells in various immune-mediated diseases. We are now setting up a system to identify various subtypes of leukocytes from peripheral blood mononuclear cells (PBMC) of heathy individuals. We expect to obtain the utmost unbiased relationship between genotypes and gene expression from healthy donors. Cell separation is performed by fluorescence-activated cell sorting into about 30 different subsets. Cells are then ana-lyzed in the steady state or in further stimulated conditions, such as with combinations of cytokines and cell surface receptor ago-nists to capture the dynamic responses of gene regulation. Firstly, genotyping as well as RNA-seq are performed. With this data, we will obtain eQTL as well as splicing QTL information. Cap analysis of gene expression (CAGE), assay for transposase-accessible chromatin using sequencing (ATAC-seq) and several histone mark analyses for each subset are powerful tools to be used for identifying the causal relationship between genetic variation and gene expression.
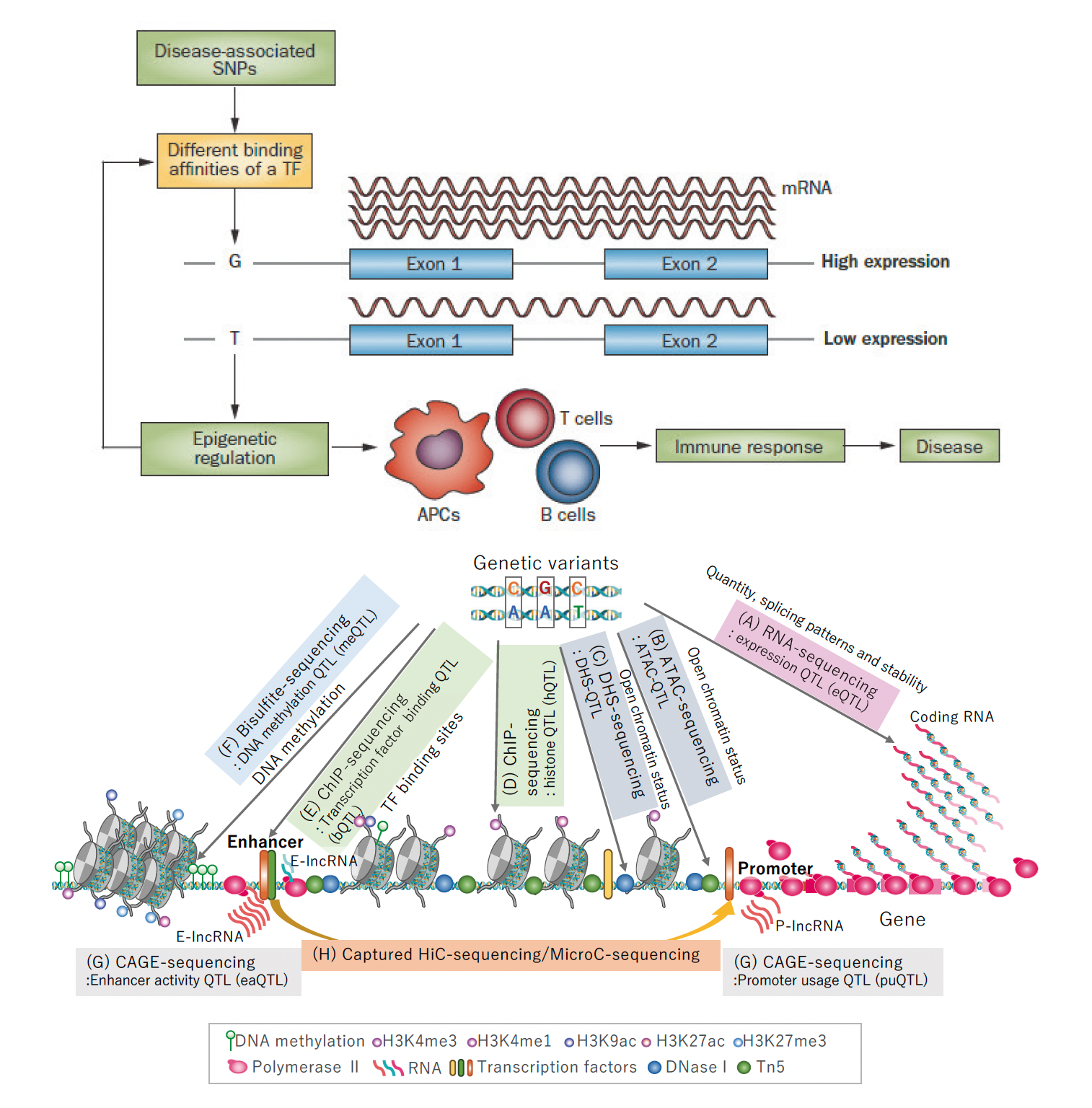
Figure: Integration of genetic information into immune functions: The eQTL project