Projects
Human genome analysis
In 2015, the Japanese government set rare hereditary diseases, cancer, dementia, infection, and pharmacogenomics as priori-ty areas for the implementation of genomic information for actual medical practice. To accomplish this goal, a combination analysis of germline variants with other information including somatic variations, gene expression profiles, and environmental factors would be key.IMS has analyzed various diseases and phenotypes by genome-wide association studies and/or targeted- and whole-genome se-quencing-based association studies, including cancer (Momozawa & Nakagawa), pharmacogenomics (Mushiroda), bone and joint diseases (Ikegawa), diabetes (Horikoshi), cardiovascular diseases (Ito), autoimmune diseases (Yamamoto K), and integrated analysis of all data and phenotypes (Terao). In addition, we began to extract information of somatic variations from DNA microarray data, which had previously been used only to call germline variants. Further, to better understand disease biology, we integrated our results with knowledge of non-coding regions and single cell se-quencing approaches by laboratories of the FANTOM and Human Cell Atlas projects. Finally, we have established collaborations with large Japanese cohorts including BioBank Japan (BBJ), Tohoku Medical Megabank, and domestic and international universities.A key finding in 2020 was elucidating the basics underlying mosaic chromosomal alterations (mCA), clonal hematopoiesis with somatic chromosomal alterations. We identified 33,250 mCAs in 179,417 BBJ participants and found that mCA seems inevitable with advancing age. Key differences in the genomic locations of mCA between Japanese and Europeans could help predict the relative rates of B-cell and T-cell leukemia between the populations.
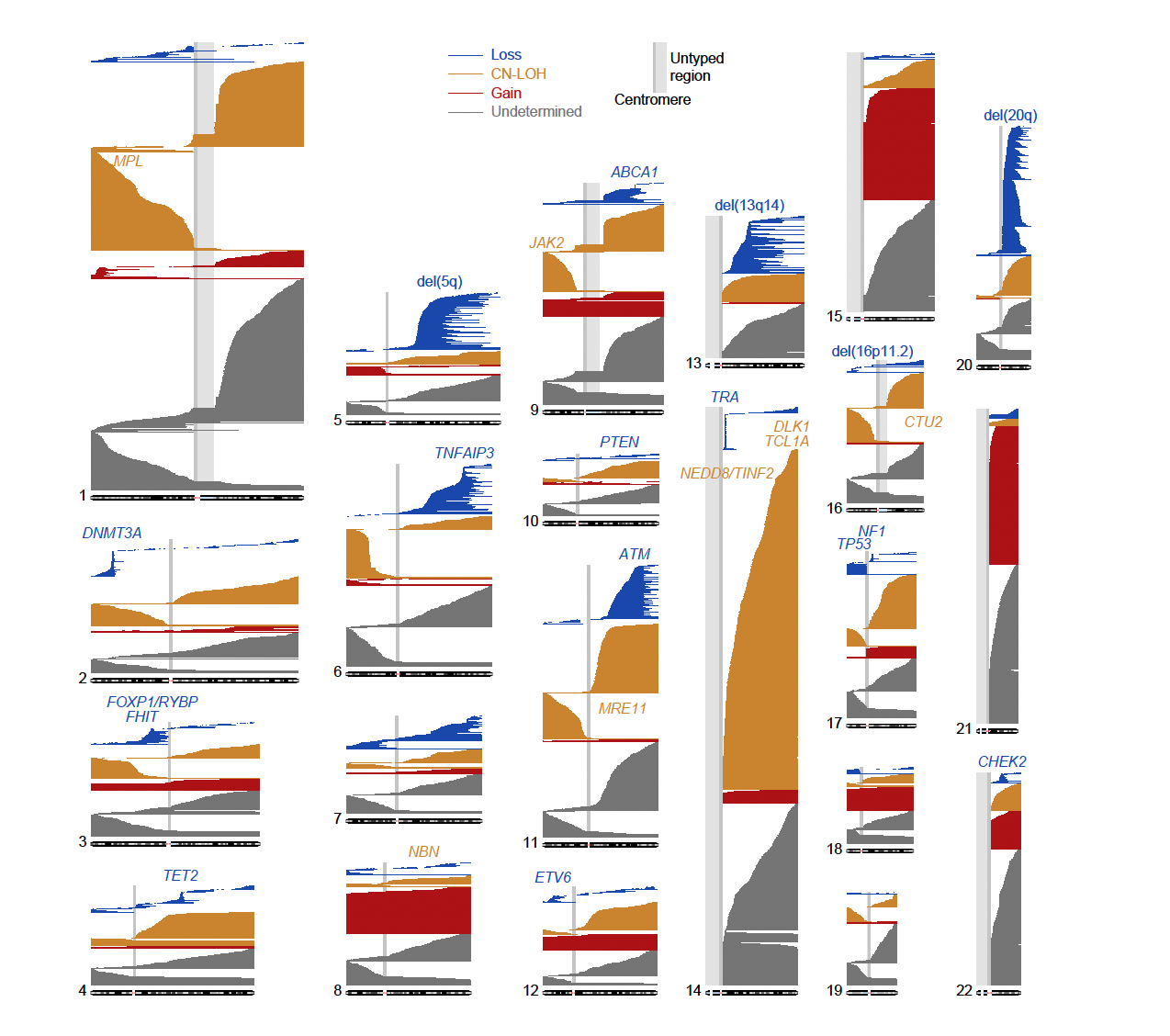
Figure: Genomic locations of 33,250 autosomal mCAs detected in 27,910 unique BBJ participants
mCA events are plotted as blue, orange and red horizontal lines, according to their mCA classes. Events with undetermined copy numbers are plotted in grey. Commonly deleted regions are labelled in blue; loci associated with CN-LOH mutations in cis are labelled in orange.